The impact of the existence or absence of CM on fish’s intestinal microbiota environment
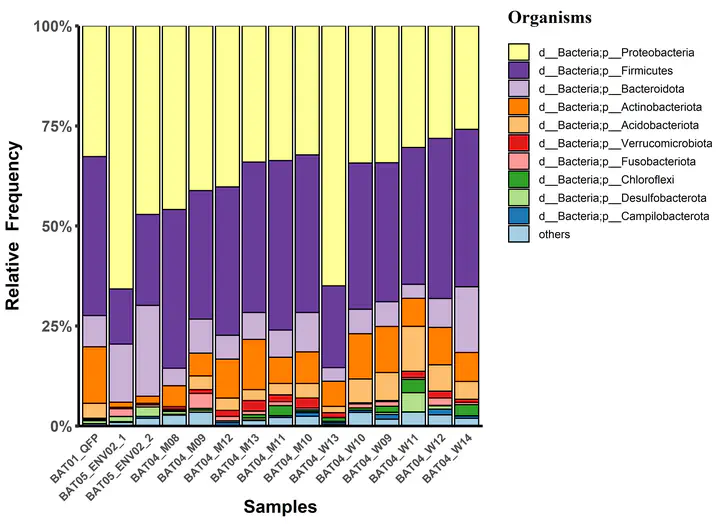 Taxonomic composition Barplot
Taxonomic composition Barplot
The chitin-based peritrophic matrix (PM) plays a crucial role in gut immunity and digestion in invertebrates. Traditionally, PM was believed to be absent in all vertebrates, however, a recently found PM-like chitinous membrane (CM) in fishes has challenged this notion and has the potential to enhance our understanding of vertebrate gut physiology and structural evolution.
In this project, our team aimed to investigate if the existence of the CM in zebrafish gut would have an benefical or harmful impact on the survival of the fish, these two may have no correlation of course. To fullfill our goal, firstly, We created a complete genetic PM/CM knockout model through the prevalent CRISPR-Cas9 gene editing technology. Then, we designed a series of rigorous experiments to test the influences of the absence of the CM on the fishes, including ingestion behavior, microbial homeostasis, epithelial renewal, digestion, growth and longevity.
As a bioinformatics Mphil candidate, I was entrusted with the mission of designing a accurate and stable pipeline to analyse the 16S amplicon sequencing data which was a complexed work obviously. The first challenge we encountered was how to select a suitable platform for conducting our data analysis as there are many microbial data analysis platforms in the field. we ultimately decided to integrate the USEARCH and QIIME 2 for our data analysis after careful comparison. Specifically, USEARCH is known for its simplicity and refinement while QIIME 2 is powerful and comprehensive, they complement each other excellently. In fact, we solely utilized USEARCH to generate the feature table, which was the central step in the entire process and subsequent analyses were primarily conducted using relevant plugins from QIIME 2. In order to enhance the reproducibility of the data analysis and facilitate potential interested researchers, I developed a simple software called “LuckyMicrobe.” Detailed information about this software can be found on my GitHub page(https://github.com/Learnerhua/LuckyMicrobe-testing- ), though it is still in the experimental stage.
Currently, this research topic is still being further extended, and our team has a continuous demand for data analysis. As a result, this project remains ongoing, and I continue to improve my software constantly.