Comparative Genomic Analysis of the ApeC Protein Family.
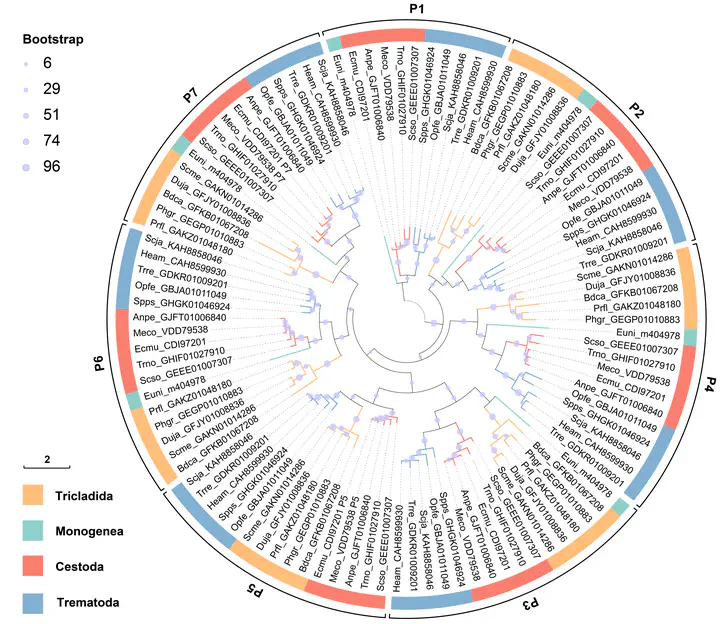 phylogenetic analysis of ACPs in Platyhelminthes
phylogenetic analysis of ACPs in Platyhelminthes
The ApeC domain was originally defined by our team in 2014 within two pattern recognition receptor(PRRs) from amphioxus, though these proteins, known as apextrins, were first discovered over twenty years ago. Generally, we refer to proteins containing the ApeC domain as ACPs. Recent studies have confirmed that ACPs show agglutinating activity but no microbicidal activity to bacteria and participate in controlling the activation of the NF-κB pathway by interacting with TRAF6 moleculer. Meanwhile, the distribution of ACPs across the taxa has also been elucidated which suggested that they are mainly distributed in invertebrates and can be traced back to early prokaryotes, such as bacteria.
Building upon the previous research, our team is dedicated to clarifying the characteristics and functions of ACPs in various phyla. In our latest research, we further revealed the distribution, structure, and functional characteristics of ACPs in the phylum Mollusca(taxonomy ID:6647). I was assigned one of the tasks involving data analysis. Simultaneously, I am taking charge of the analysing work of ACPs in the phylum platyhelminthes(taxonomy ID: 6157), normally known as the flatworm, the species within this phylum are mostly famous for their unparalleled regenerative abilities or notorious parasitic characteristics. Given the fact that intensive aquaculture is severely impacted by parasites, particularly monogenean, our research has been endowed with more practical meaning.
Up to now, I have successfully accomplished the majority of tasks in the project and achieved numerous significant research outcomes, including assuring the existence of ACPs in Monogenean and free-living flatworms for the first time, finding that the ApeC domains in flatworms exhibit a repetitive tandem arrangement and so on. I am currently organizing these findings into a publishable academic paper and plan to publish it in the near future.
Meanwhile, I have not halted my research efforts and am collaborating with others to explore the functional patterns of ACPs in bacteria.